Microbiology studies individual microorganisms through cultivation and microscopy, enabling detailed analysis of their physiology and genetics. Metagenomics analyzes genetic material directly from environmental samples, providing insights into complex microbial communities without the need for culturing. This approach reveals the diversity and functional potential of microbes in various habitats, complementing traditional microbiology techniques.
Table of Comparison
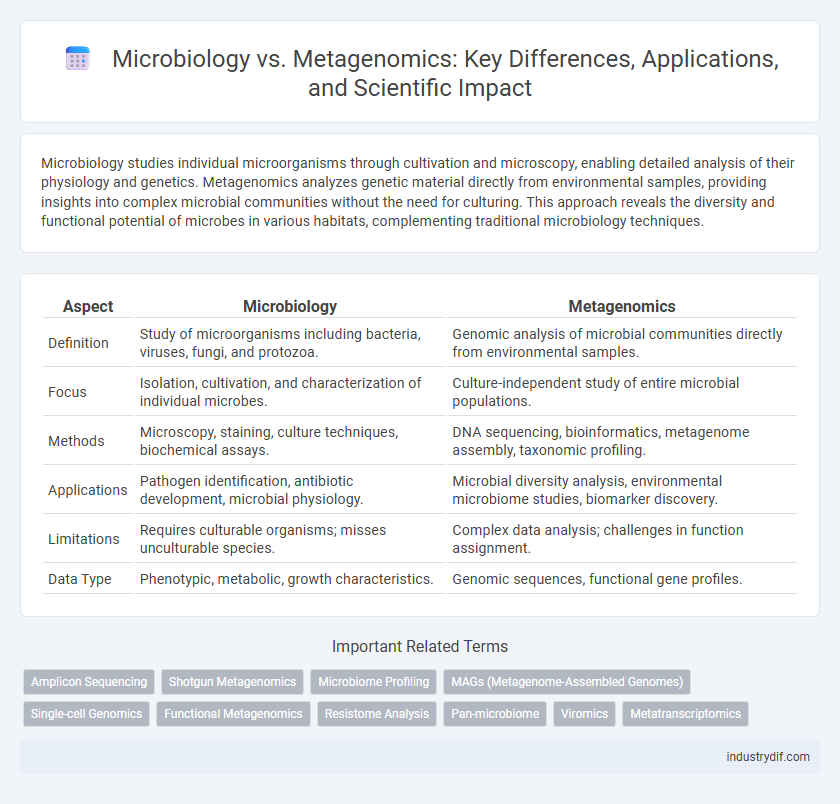
| Aspect | Microbiology | Metagenomics |
|---|---|---|
| Definition | Study of microorganisms including bacteria, viruses, fungi, and protozoa. | Genomic analysis of microbial communities directly from environmental samples. |
| Focus | Isolation, cultivation, and characterization of individual microbes. | Culture-independent study of entire microbial populations. |
| Methods | Microscopy, staining, culture techniques, biochemical assays. | DNA sequencing, bioinformatics, metagenome assembly, taxonomic profiling. |
| Applications | Pathogen identification, antibiotic development, microbial physiology. | Microbial diversity analysis, environmental microbiome studies, biomarker discovery. |
| Limitations | Requires culturable organisms; misses unculturable species. | Complex data analysis; challenges in function assignment. |
| Data Type | Phenotypic, metabolic, growth characteristics. | Genomic sequences, functional gene profiles. |
Defining Microbiology and Metagenomics
Microbiology is the scientific study of microorganisms, including bacteria, viruses, fungi, and protozoa, focusing on their physiology, genetics, and interactions with environments and hosts. Metagenomics involves the analysis of genetic material recovered directly from environmental samples, enabling the study of microbial communities without the need for culturing individual species. This approach provides comprehensive insights into microbial diversity, function, and ecology at a community level.
Historical Evolution of Microbiology and Metagenomics
Microbiology, established in the 17th century with Antonie van Leeuwenhoek's invention of the microscope, laid the foundation for studying microorganisms through culture-based techniques. The emergence of metagenomics in the early 21st century revolutionized microbial research by enabling DNA sequencing directly from environmental samples, bypassing traditional culturing limitations. This shift has accelerated discoveries in microbial diversity, ecology, and function by providing comprehensive insights into complex microbial communities.
Core Methodologies: Culturing vs Sequencing
Microbiology primarily relies on culturing techniques to isolate and grow microorganisms under controlled laboratory conditions, enabling detailed phenotypic and biochemical analyses. Metagenomics employs high-throughput sequencing of environmental DNA samples, bypassing the need for culturing and allowing comprehensive identification of microbial communities, including unculturable species. These core methodologies contrast in their approach, with culturing offering specificity and functional insights, while sequencing provides extensive biodiversity and genomic information.
Microbial Diversity: Limitations and Opportunities
Microbiology traditionally relies on culturing techniques that capture only a fraction of microbial diversity, often missing unculturable or rare species. Metagenomics offers a culture-independent approach, enabling comprehensive analysis of microbial communities through direct sequencing of environmental DNA. Despite challenges like data complexity and incomplete reference databases, metagenomics significantly expands opportunities for discovering novel microbes and understanding ecosystem functions.
Applications in Healthcare and Environmental Science
Microbiology enables the isolation and characterization of individual microorganisms to diagnose infections and develop targeted antibiotics in healthcare, while metagenomics offers comprehensive analysis of microbial communities, enhancing disease surveillance and antibiotic resistance tracking. In environmental science, microbiology supports bioremediation efforts by studying specific pollutant-degrading bacteria, whereas metagenomics facilitates ecosystem monitoring through the assessment of microbial diversity and functional potential in various habitats. Both approaches are critical for advancing precision medicine and sustainable environmental management by leveraging microbial data at different scales.
Data Analysis: Bioinformatics in Microbiology and Metagenomics
Bioinformatics plays a critical role in microbiology by enabling the analysis of genomic sequences from individual microbial isolates, focusing on gene annotation, phylogenetics, and metabolic pathway reconstruction. In metagenomics, bioinformatics tools process massive, complex datasets derived from environmental samples, facilitating taxonomic profiling, functional annotation, and diversity assessment without the need for culturing organisms. Advanced algorithms and databases such as QIIME, MEGAHIT, and MG-RAST generate insights into microbial community structure and function, driving discoveries in ecosystem dynamics and disease ecology.
Challenges in Microbial Identification and Classification
Microbiology faces challenges in microbial identification due to the limitations of culturing techniques, which exclude many unculturable microorganisms from analysis. Metagenomics overcomes these barriers by enabling culture-independent sequencing of genetic material directly from environmental samples, revealing vastly greater microbial diversity. However, metagenomic data interpretation is complicated by incomplete reference databases and the complexity of distinguishing closely related microbial species.
Technological Advances Driving Both Fields
High-throughput sequencing technologies have revolutionized microbiology by enabling precise identification and characterization of individual microbial species, while metagenomics leverages these advances to analyze complex microbial communities without cultivation. Innovations such as single-cell genomics and long-read sequencing enhance resolution in microbiology and facilitate comprehensive insights into community structure and function in metagenomics. Bioinformatics tools optimized for large-scale data integration and functional annotation drive progress in both fields, expanding understanding of microbial diversity and ecosystem dynamics.
Integration of Microbiology and Metagenomics in Research
Integration of microbiology and metagenomics enhances the understanding of microbial communities by combining traditional cultivation techniques with high-throughput DNA sequencing. This approach enables comprehensive analysis of microbial diversity, gene function, and metabolic pathways in complex environments without the need for culturing individual species. Leveraging metagenomic data alongside microbiological methods accelerates discoveries in microbial ecology, disease mechanisms, and biotechnological applications.
Future Directions in Microbial Science
Future directions in microbial science emphasize integrating metagenomics with traditional microbiology to uncover the vast genetic diversity of uncultured microorganisms. Advances in high-throughput sequencing and bioinformatics enable detailed functional profiling of microbial communities, providing insights into their ecological roles and potential biotechnological applications. These approaches accelerate discoveries in antibiotic resistance, microbial interactions, and ecosystem dynamics, revolutionizing diagnostics and environmental management.
Related Important Terms
Amplicon Sequencing
Amplicon sequencing, a targeted approach within metagenomics, enables high-resolution profiling of microbial communities by selectively amplifying and sequencing specific genetic markers such as the 16S rRNA gene. This method contrasts with traditional microbiology techniques, which often rely on culturing, by allowing comprehensive analysis of microbial diversity, abundance, and functional potential without the need for cultivation.
Shotgun Metagenomics
Shotgun metagenomics enables comprehensive analysis of microbial communities by sequencing all genetic material directly from environmental samples, unlike traditional microbiology which relies on culturing individual species. This high-throughput approach reveals functional potential and diversity, offering deeper insights into complex microbiomes beyond the limitations of culture-dependent methods.
Microbiome Profiling
Microbiology employs culture-based techniques and targeted molecular assays for microbiome profiling, enabling characterization of specific microbial taxa and their functions. Metagenomics leverages high-throughput sequencing to analyze entire microbial communities' genetic material, providing comprehensive insights into microbiome diversity and functional potential without the need for culturing.
MAGs (Metagenome-Assembled Genomes)
Microbiology traditionally relies on culturing techniques to study individual microbial species, while metagenomics enables the reconstruction of Metagenome-Assembled Genomes (MAGs) directly from environmental samples, bypassing the need for cultivation. MAGs provide comprehensive insights into microbial diversity, functional potential, and genome organization within complex communities, revolutionizing microbial ecology and evolution studies.
Single-cell Genomics
Single-cell genomics enables the analysis of individual microbial cells, providing high-resolution insights into microbial diversity and function that traditional microbiology and bulk metagenomics techniques may overlook. This approach enhances understanding of microbial heterogeneity, gene expression, and metabolic pathways at the single-cell level, facilitating breakthroughs in environmental microbiology and infectious disease research.
Functional Metagenomics
Functional metagenomics enables the direct analysis of microbial gene functions from environmental samples without the need for culturing, making it a powerful tool to discover novel enzymes and bioactive compounds. Unlike traditional microbiology, which studies isolated microbes, functional metagenomics leverages high-throughput sequencing and gene expression assays to uncover microbial diversity and metabolic potential within complex communities.
Resistome Analysis
Resistome analysis in microbiology involves characterizing antibiotic resistance genes within cultured microbial isolates, whereas metagenomics enables comprehensive profiling of resistomes directly from environmental or clinical samples without the need for cultivation. Metagenomic approaches reveal the diversity and distribution of resistance genes across microbial communities, facilitating the identification of novel resistance determinants and tracking of resistome dynamics in complex ecosystems.
Pan-microbiome
Pan-microbiome analysis extends traditional microbiology by integrating metagenomics to capture the full spectrum of microbial diversity across multiple environments and hosts, revealing collective genetic potential beyond isolated microbial communities. This approach enhances understanding of microbial interactions, functional capabilities, and evolutionary dynamics within complex ecosystems, offering critical insights into microbiome variability and resilience.
Viromics
Viromics, a sub-discipline of metagenomics, employs high-throughput sequencing to analyze viral communities within environmental samples, providing comprehensive insights into viral diversity and function beyond traditional microbiology's isolate-based methods. Unlike conventional microbiology that focuses on culturable viruses, viromics enables the study of unculturable and novel viruses, enhancing understanding of viral ecology, evolution, and their roles in microbial ecosystems.
Metatranscriptomics
Metatranscriptomics analyzes the active gene expression of microbial communities by sequencing RNA transcripts, providing insights into functional activity and dynamic responses that traditional microbiology and metagenomics, which focus on microbial identification and genomic potential respectively, cannot capture. This approach enables the study of metabolic pathways and microbial interactions in situ, revealing real-time physiological states and environmental adaptations of microorganisms.
Microbiology vs Metagenomics Infographic
 industrydif.com
industrydif.com