Genomics involves sequencing and analyzing the entire genetic material of a single organism to understand its structure, function, and evolution. Pangenomics expands this approach by comparing the genomes of multiple individuals within a species to identify shared and unique genes, capturing genetic diversity and adaptation mechanisms. This broader perspective enhances insights into population variation, disease resistance, and evolutionary biology.
Table of Comparison
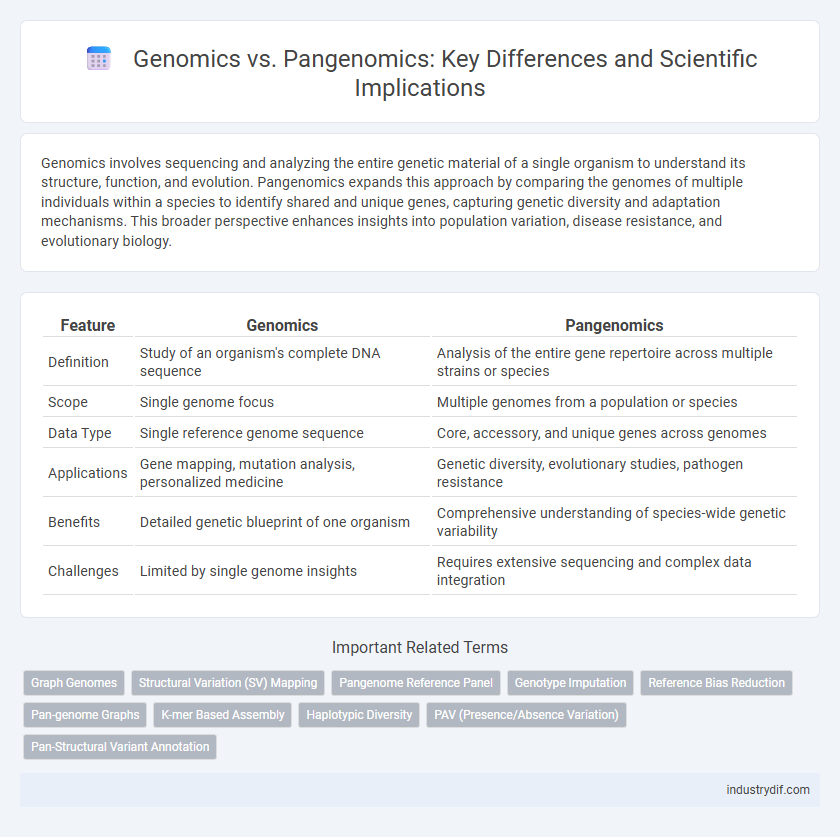
| Feature | Genomics | Pangenomics |
|---|---|---|
| Definition | Study of an organism's complete DNA sequence | Analysis of the entire gene repertoire across multiple strains or species |
| Scope | Single genome focus | Multiple genomes from a population or species |
| Data Type | Single reference genome sequence | Core, accessory, and unique genes across genomes |
| Applications | Gene mapping, mutation analysis, personalized medicine | Genetic diversity, evolutionary studies, pathogen resistance |
| Benefits | Detailed genetic blueprint of one organism | Comprehensive understanding of species-wide genetic variability |
| Challenges | Limited by single genome insights | Requires extensive sequencing and complex data integration |
Defining Genomics and Pangenomics
Genomics is the comprehensive study of an organism's complete DNA sequence, including all of its genes and their functions, variations, and interactions within a single genome. Pangenomics expands this concept by analyzing the entire gene repertoire across multiple strains or species, capturing both core genes shared by all and accessory genes unique to subsets, revealing genetic diversity and evolutionary dynamics. This comparative approach enables identifying novel genetic elements and understanding population-level variations beyond individual genomes.
Historical Evolution of Genomics and Pangenomics
Genomics originated in the late 20th century with the sequencing of entire genomes, surpassing traditional gene-by-gene analysis and enabling comprehensive studies of individual organisms. Pangenomics emerged in the 21st century as a response to genomic diversity, capturing the full complement of genes within a species across multiple strains or individuals, which expanded understanding beyond a single reference genome. The evolution from genomics to pangenomics reflects advancements in high-throughput sequencing technologies and computational methods that facilitate comparative genomic analysis at a population level.
Methodological Approaches in Genomics
Genomics primarily utilizes whole-genome sequencing and comparative analysis to study the genetic material of a single organism, focusing on reference genomes and variant detection. Pangenomics extends this approach by integrating multiple genomes from a species or population to capture genomic diversity, employing graph-based models and multi-genome alignments for comprehensive representation. Methodological advances in genomics such as long-read sequencing, high-throughput data generation, and machine learning algorithms enable detailed annotation and functional interpretation that support pangenomic frameworks.
Advances in Pangenomic Techniques
Recent advances in pangenomic techniques, such as high-throughput sequencing and graph-based genome assembly, enable comprehensive analysis of genetic diversity beyond single reference genomes. These methods capture structural variations, presence-absence variations, and sequence polymorphisms across multiple individuals or species, providing deeper insights into evolutionary dynamics and functional genomics. Integrating long-read sequencing technologies and machine learning algorithms further enhances accuracy and resolution in constructing complex pangenomes.
Comparative Analysis: Genomics vs Pangenomics
Genomics examines the complete DNA sequence of a single organism, identifying genes and genetic variations within that individual. Pangenomics expands this approach by analyzing the collective genome of multiple strains or species, capturing core and accessory genes to understand genetic diversity. Comparative analysis reveals that pangenomics provides a broader perspective on population-level genetic variations, while traditional genomics offers detailed insights into single-genome structure and function.
Applications in Precision Medicine
Genomics analyzes individual genomes to identify specific genetic variants linked to disease, enabling targeted therapies and personalized treatment plans. Pangenomics expands this approach by capturing genetic diversity across populations, improving the accuracy of variant interpretation and the development of inclusive precision medicine strategies. Integrating pangenomic data enhances the prediction of drug response and disease susceptibility across diverse genetic backgrounds, optimizing patient outcomes.
Impact on Genetic Diversity Studies
Genomics analyzes the complete DNA sequence of a single organism, providing detailed insight into its genetic makeup and mutation patterns. Pangenomics expands this approach by comparing multiple genomes within a species, capturing core and accessory genes that reveal broader genetic diversity and evolutionary dynamics. This comparative framework enhances understanding of population structure, adaptability, and functional variability crucial for studying genetic diversity across species.
Data Integration and Computational Challenges
Genomics focuses on analyzing the complete DNA sequence of an individual organism, enabling insights into genetic variations and functional elements within a single genome. Pangenomics extends this approach by integrating multiple genomes from a species or population, capturing structural variations and gene presence/absence that single reference genomes often miss. The computational challenges in pangenomics involve managing and interpreting vast, complex datasets requiring advanced algorithms and scalable storage solutions to efficiently process diverse genomic architectures.
Future Trends in Genomic and Pangenomic Research
Future trends in genomic and pangenomic research emphasize integrating multi-omics data to enhance precision medicine and biodiversity conservation. Advances in AI-driven algorithms and high-throughput sequencing technologies accelerate the identification of novel genetic variants and complex traits across diverse populations. Embracing pangenome frameworks enables comprehensive representation of genomic diversity, facilitating breakthroughs in evolutionary biology and personalized therapeutics.
Ethical Considerations in Genome Science
Ethical considerations in genomics and pangenomics center on data privacy, informed consent, and equitable access to genetic information. Genomics typically involves individual genome analysis, raising concerns about personal data security and potential misuse of sensitive health information. Pangenomics, which examines the collective genetic diversity across populations, introduces challenges in representing diverse groups fairly and addressing potential biases in genomic databases.
Related Important Terms
Graph Genomes
Graph genomes in pangenomics represent a revolutionary approach by capturing genomic diversity through directed acyclic graphs that integrate multiple individual genomes, enabling more comprehensive variant detection than traditional linear reference genomics. This method enhances the resolution of complex structural variations, improving the accuracy of genotyping and fostering deeper insights into population genetics and disease associations.
Structural Variation (SV) Mapping
Structural variation mapping in genomics identifies large-scale DNA alterations within a single reference genome, while pangenomics captures a comprehensive catalog of SVs across multiple individuals, enhancing detection of population-level genetic diversity. Advanced pangenomic approaches improve SV resolution by integrating diverse haplotypes, enabling more accurate association studies and evolutionary analyses.
Pangenome Reference Panel
Pangenome reference panels map the full genomic diversity within a species by integrating multiple individual genomes, surpassing traditional single-reference genomics that often miss structural variants and rare alleles. This comprehensive approach enables more accurate variant detection, enhances population genetics studies, and supports precision medicine by reflecting a broader spectrum of genetic variation.
Genotype Imputation
Genotype imputation enhances genomic studies by predicting unobserved genotypes using reference panels, improving variant density beyond direct genotyping arrays. Pangenomics extends this approach by incorporating diverse population genomes, creating a comprehensive reference that increases imputation accuracy and captures structural variation missed in single-reference genomic analyses.
Reference Bias Reduction
Pangenomics expands beyond traditional genomics by incorporating multiple genomes within a species, significantly reducing reference bias inherent in single-reference analyses. This inclusive approach enhances the accuracy of variant detection and evolutionary studies by representing the full spectrum of genetic diversity.
Pan-genome Graphs
Pan-genome graphs represent genomic diversity by integrating multiple individual genomes into a single, comprehensive structure, enabling detailed analysis of genetic variation beyond linear reference genomes. This graph-based approach facilitates more accurate variant detection and functional annotation, advancing personalized medicine and evolutionary studies in genomics and pangenomics.
K-mer Based Assembly
K-mer based assembly in genomics involves breaking down sequences into fixed-length substrings to reconstruct genomes, enabling efficient handling of repetitive regions and sequencing errors. In contrast, pangenomics leverages k-mer assemblies across multiple genomes to capture pan-genomic diversity, identifying core and accessory genes for comprehensive population-level analysis.
Haplotypic Diversity
Genomics analyzes individual genomes to identify genetic variations, while pangenomics integrates multiple genomes to capture the full spectrum of haplotypic diversity across populations. This comprehensive approach enhances the understanding of allele combinations and structural variants crucial for studying genetic traits and disease susceptibility.
PAV (Presence/Absence Variation)
Presence/Absence Variation (PAV) highlights genomic differences by capturing gene content variability across multiple genomes, a key focus in pangenomics that extends beyond single reference genomes analyzed in traditional genomics. Pangenomics enables comprehensive mapping of PAV to reveal structural variations and gene gain/loss events critical for understanding species diversity and evolution.
Pan-Structural Variant Annotation
Pan-structural variant annotation in pangenomics enables comprehensive identification of genomic variations by comparing multiple genomes within a species, surpassing traditional genomics that relies on a single reference genome. This approach enhances the detection of complex structural variants critical for understanding genetic diversity, evolution, and disease associations.
Genomics vs Pangenomics Infographic
 industrydif.com
industrydif.com