PCR machines amplify DNA segments through repeated thermal cycling, providing qualitative or semi-quantitative data useful in various molecular biology applications. Digital PCR offers absolute quantification by partitioning samples into thousands of micro-reactions, enhancing sensitivity and precision for detecting low-abundance targets or rare mutations. While traditional PCR is faster and more cost-effective for routine analysis, digital PCR excels in applications requiring high accuracy and detailed quantification.
Table of Comparison
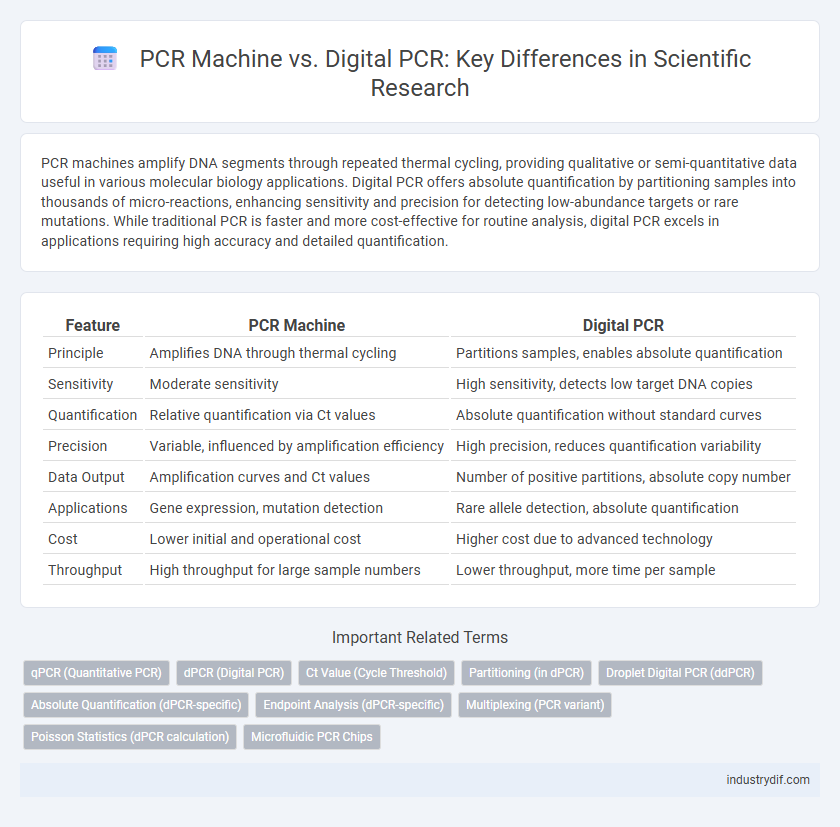
| Feature | PCR Machine | Digital PCR |
|---|---|---|
| Principle | Amplifies DNA through thermal cycling | Partitions samples, enables absolute quantification |
| Sensitivity | Moderate sensitivity | High sensitivity, detects low target DNA copies |
| Quantification | Relative quantification via Ct values | Absolute quantification without standard curves |
| Precision | Variable, influenced by amplification efficiency | High precision, reduces quantification variability |
| Data Output | Amplification curves and Ct values | Number of positive partitions, absolute copy number |
| Applications | Gene expression, mutation detection | Rare allele detection, absolute quantification |
| Cost | Lower initial and operational cost | Higher cost due to advanced technology |
| Throughput | High throughput for large sample numbers | Lower throughput, more time per sample |
Introduction to PCR Machine and Digital PCR
PCR machines utilize thermal cycling to amplify DNA by repeatedly denaturing, annealing, and extending target sequences, enabling rapid and exponential amplification of genetic material. Digital PCR partitions a DNA sample into thousands of individual reactions, allowing precise quantification by counting positive amplification events, enhancing sensitivity and accuracy in detecting low-abundance targets. Both technologies are essential in molecular biology, but digital PCR offers superior quantification capabilities compared to traditional PCR machines.
Principles of Conventional PCR vs Digital PCR
Conventional PCR amplifies DNA by cycling through denaturation, annealing, and extension phases, producing exponential amounts of target sequences measured through bulk fluorescence. Digital PCR partitions the sample into thousands of micro-reactions, allowing absolute quantification of nucleic acids based on the presence or absence of amplification in each partition. This partitioning enhances sensitivity and precision by enabling direct counting of target molecules without the need for standard curves.
Key Differences Between PCR Machine and Digital PCR
PCR machines amplify DNA through cyclical temperature changes enabling exponential DNA replication, while digital PCR partitions the sample into thousands of micro-reactions for absolute quantification. Digital PCR offers higher sensitivity, precision, and quantification accuracy compared to conventional PCR, making it ideal for detecting rare mutations and low-abundance targets. The primary difference lies in the output format: PCR produces relative quantification via amplification curves, whereas digital PCR provides absolute quantification by direct counting of DNA molecules.
Sensitivity and Accuracy: PCR vs Digital PCR
Digital PCR offers superior sensitivity and accuracy compared to traditional PCR by enabling absolute quantification of nucleic acids without the need for standard curves. The partitioning of samples into thousands of micro-reactions in digital PCR reduces background noise and allows detection of low-abundance targets with high precision. Traditional PCR relies on amplification cycles and fluorescence thresholds, which can introduce variability and limit accuracy in detecting rare DNA sequences.
Quantification Capabilities: Traditional vs Digital
Traditional PCR machines provide relative quantification by amplifying DNA and measuring fluorescence at each cycle, but this method relies heavily on calibration curves and exponential amplification efficiency. Digital PCR (dPCR) partitions the sample into thousands of individual reactions, allowing absolute quantification of nucleic acid copies without the need for standard curves. The high sensitivity and precision of dPCR enable accurate detection of low-abundance targets and subtle fold changes in gene expression, surpassing the quantification capabilities of conventional PCR.
Workflow and Operation Differences
PCR machines rely on traditional thermal cycling to amplify DNA, requiring precise temperature changes through denaturation, annealing, and extension phases. Digital PCR partitions the sample into thousands of micro-reactions, enabling absolute quantification by counting positive reactions with increased sensitivity and precision. Workflow in digital PCR involves sample partitioning and complex data analysis, whereas conventional PCR emphasizes rapid cycling and endpoint detection.
Applications in Research and Diagnostics
PCR machines enable amplification of DNA sequences for genetic research, pathogen detection, and mutation analysis, providing qualitative and quantitative data. Digital PCR offers higher precision by partitioning samples into thousands of reactions, enhancing sensitivity in rare allele detection, viral load measurement, and copy number variation studies. Both technologies facilitate advancements in cancer research, infectious disease diagnostics, and personalized medicine through accurate nucleic acid quantification.
Cost and Accessibility Comparison
PCR machines typically offer lower initial costs and greater accessibility due to their widespread availability and established technology, making them suitable for many standard laboratory applications. Digital PCR systems require higher upfront investment and specialized training, limiting accessibility to advanced research facilities and clinical laboratories. However, the precise quantification and sensitivity of Digital PCR justify the increased cost in applications requiring high accuracy.
Limitations and Challenges of Each Method
PCR machines face limitations such as susceptibility to contaminants and inefficient quantification at low DNA concentrations, while digital PCR offers higher sensitivity but is constrained by higher costs and complexity in data interpretation. Digital PCR requires precise partitioning of the sample and can suffer from issues related to droplet stability and throughput limitations. Both methods encounter challenges in scaling for large sample sizes and maintaining reproducibility across different laboratories.
Future Trends in PCR Technology
Future trends in PCR technology highlight the increasing adoption of digital PCR (dPCR) due to its superior precision in quantifying nucleic acids and enhanced sensitivity for detecting low-abundance targets. Innovations focus on integrating microfluidics and automation to improve throughput and reduce contamination risks in both traditional PCR machines and dPCR platforms. Emerging approaches also emphasize multiplexing capabilities and real-time data analytics powered by artificial intelligence to accelerate diagnostics and personalized medicine applications.
Related Important Terms
qPCR (Quantitative PCR)
qPCR (Quantitative PCR) amplifies and quantifies DNA by measuring fluorescence during each PCR cycle, enabling precise gene expression analysis and pathogen detection. Digital PCR partitions the sample into thousands of micro-reactions, offering absolute quantification without the need for standard curves and higher sensitivity for rare target detection.
dPCR (Digital PCR)
Digital PCR (dPCR) offers precise quantification of nucleic acids by partitioning samples into thousands of micro-reactions, significantly enhancing sensitivity and accuracy compared to conventional PCR machines. Its ability to detect low-abundance mutations and absolute DNA copy number without standard curves makes dPCR essential for applications in diagnostics, oncology, and pathogen detection.
Ct Value (Cycle Threshold)
PCR machines generate Ct values by amplifying target DNA sequences through multiple cycles, with the Ct value indicating the cycle number at which fluorescence surpasses the threshold, reflecting initial DNA quantity. Digital PCR offers enhanced precision by partitioning samples into numerous micro-reactions, enabling absolute quantification without relying solely on Ct values and reducing variability caused by amplification efficiency.
Partitioning (in dPCR)
Digital PCR (dPCR) enhances sensitivity and accuracy by partitioning a sample into thousands of nanoliter-sized reactions, enabling absolute quantification of nucleic acids without reference standards. This partitioning step contrasts with traditional PCR machines, which amplify target sequences in bulk reactions, limiting detection of low-abundance variants and mixed populations.
Droplet Digital PCR (ddPCR)
PCR machines enable DNA amplification through thermal cycling, while Droplet Digital PCR (ddPCR) enhances quantification accuracy by partitioning samples into thousands of nanoliter droplets, allowing absolute DNA copy number determination without standard curves. ddPCR demonstrates superior sensitivity and precision compared to conventional PCR, making it essential for detecting rare mutations, low abundance targets, and complex genomic variations in scientific research.
Absolute Quantification (dPCR-specific)
Digital PCR (dPCR) offers highly precise absolute quantification by partitioning the sample into thousands of individual reactions, allowing direct counting of target DNA molecules without reliance on standard curves used in conventional PCR machines. This method enhances sensitivity and accuracy in detecting low-abundance nucleic acids, making it superior for applications requiring exact quantification.
Endpoint Analysis (dPCR-specific)
Digital PCR (dPCR) offers enhanced endpoint analysis by partitioning samples into thousands of individual reactions, enabling absolute quantification of nucleic acids without the need for standard curves. Unlike conventional PCR, dPCR's endpoint fluorescence measurement provides higher precision and sensitivity in detecting low-abundance targets and rare mutations.
Multiplexing (PCR variant)
Digital PCR (dPCR) offers superior multiplexing capabilities compared to traditional PCR machines, enabling simultaneous detection and quantification of multiple target sequences with higher sensitivity and precision. This enhanced multiplexing efficiency in dPCR facilitates complex genetic analyses, such as pathogen detection and gene expression profiling, by reducing cross-reactivity and improving quantification accuracy.
Poisson Statistics (dPCR calculation)
Digital PCR utilizes Poisson statistics to precisely quantify nucleic acid molecules by partitioning samples into thousands of micro-reactions, enabling absolute quantification without standard curves, unlike traditional PCR machines that rely on relative amplification measurements. This statistical approach in dPCR accounts for the probability of target molecule distribution, reducing quantification errors and improving sensitivity in detecting low-abundance genetic variants.
Microfluidic PCR Chips
Microfluidic PCR chips enhance digital PCR by enabling precise partitioning of samples into thousands of nanoliter-scale reactions, significantly improving sensitivity and quantification accuracy compared to conventional PCR machines. This technology leverages microfluidic channels to control fluid dynamics, reducing reagent consumption and accelerating thermal cycling for rapid and high-throughput nucleic acid analysis.
PCR Machine vs Digital PCR Infographic
 industrydif.com
industrydif.com